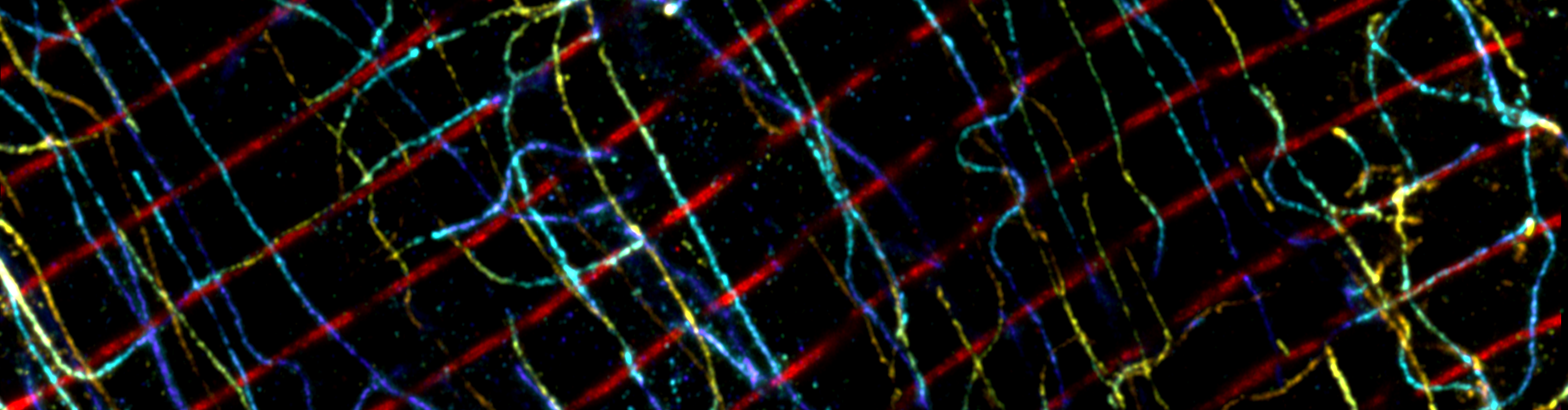
Expansion microscopy innovations and applications
Wednesday 13 May 2020
I’ve been meaning to publish this blog post on innovations in expansion microscopy for a while, and due to virus-enforced time out of the lab, I’ve now got the perfect opportunity.
I wanted to paint a picture of the plethora of ExM approaches that have appeared in the literature since the original technique’s inception in 2015. The field is moving at some speed, with many groups altering gel chemistry to improve the expansion factor and isotropy, as well as combining ExM with other imaging modalities to push the resolution-increase further. Indeed, as I continued to put off writing this post over 2 months, I had more and more things to include.
Expansion microscopy (ExM) [1] is a super-resolution imaging technique that any scientist can employ. It is a chemical-based sample preparation technique, where a fluorescently labelled sample is expanded within an acrylamide-based hydrogel. Through physical expansion, ExM moves apart fluorophores which otherwise would have been too close to discern as separate points of light. ExM enhances the effective resolving power of a microscope, enabling the user to visualise structures beneath the resolution limit that would otherwise be indistinguishable.

Various innovations have been formulated which modify the methodology and gel chemistry. proExM, arguably the most prevalent form of ExM, introduced the anchoring reagent acryloyl-X (AcX), which made the 4-fold expanding gel recipe of ExM truly accessible by allowing the use of conventional antibodies [2]. An alternative approach uses MA-NHS to anchor targets [3]. More recently, click chemistry has been employed as a strategy to link reporter molecules to the gel [4,5].
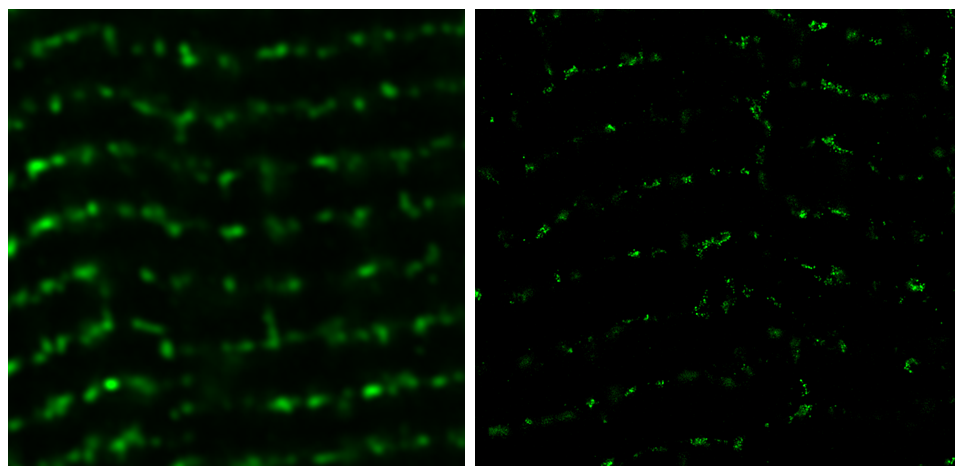
Enhanced expansion factors, leading to greater improvements in resolution can be achieved through iterative ExM (iExM), involving sequential expansion steps [6,7]. Alternatively, X10 microscopy implements the self-crosslinking dimethylacrylamide monomer, enabling 10-fold expansion in a single gel [8].
Other innovations have sought to address isotropy, the measure of minimising introduced errors. Synthetic monomers and click-chemistry have been used to obtain a more homogenous gel matrix via non-radical polymerisation [9]. U-ExM features a modified crosslinking strategy in order to preserve the target macromolecule ultrastructure [10], and similarly, differential ExM (DiExM) optimised fixation and expansion steps [11]. Origami nanorulers can be used to directly assess expansion factor at the nanoscale and gauge isotropy [12].
ExM samples have been imaged with other super-resolution imaging modalities to yield doubly improved resolution. This is no mean feat, as expanded gels are prone to drift, are compatible with only a subset of particular fluorophores, and have a different refractive index to most samples. Combinations of 4x ExM now exist with STED [13,14], SIM [15–17], STORM [18–20], SOFI [21], and light sheet microscopy [22–25]. ExM has also been combined with SABER to boost signal and allow advanced multiplexing [26]. The 10X ExM method has been combined with Airyscan microscopy [27].
A number of ExM variants have been devised which are optimised for particular applications and cell types. ExFISH enables the visualisation of RNA through the use of a small molecule linker [28]. ExM has been made compatible with lipid membrane targets [29,30], which were typically not incorporated into gels due to lacking amine groups. Cells with sturdy physical properties typically don’t fare well in the expansion process, but approaches have been devised. For instance, for the plant cell wall, digesting enzymes (cellulase, pectinase, macerozyme) have been included in the digestion step [31]. Similarly, μExM allows ExM of bacterial cells by using lysozyme and mutanolysin to break down the peptidoglycan cell wall [32]. One of the more recent developments is ExM designed for use in the model organism C. elegans, called ExCel [33]. ExPath allows the expansion of paraffin-embedded clinical tissue specimens, by a series of xylene and ethanol steps to remove the paraffin which would otherwise be detrimental to gel penetration and expansion [34].
This is by no means all the innovative uses of ExM, but I hope conveys the breadth of scope of it’s current use in scientific research.
1. Chen, F., Tillberg, P. W. & Boyden, E. S. Expansion microscopy. Science 347, 543–548 (2015).
2. Tillberg, P. W. et al. Protein-retention expansion microscopy of cells and tissues labeled using
standard fluorescent proteins and antibodies. Nat Biotechnol 34, 987–92 (2016).
3. Chozinski, T. J. et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat Methods 13, (2016).
4. Sun, D. et al. Click-ExM enables expansion microscopy for all biomolecules. bioRxiv 2020.03.19.998039 (2020) doi:10.1101/2020.03.19.998039.
5. Wen, G. et al. Evaluation of Direct Grafting Strategies via Trivalent Anchoring for Enabling Lipid Membrane and Cytoskeleton Staining in Expansion Microscopy. ACS Nano (2020) doi:10.1021/acsnano.9b09259.
6. Chang, J.-B. et al. Iterative expansion microscopy. Nat Meth advance online publication, (2017).
7. M’Saad, O. & Bewersdorf, J. Light microscopy of proteins in their ultrastructural context. bioRxiv 2020.03.13.989756 (2020) doi:10.1101/2020.03.13.989756.
8. Truckenbrodt, S. et al. X10 expansion microscopy enables 25‐nm resolution on conventional microscopes. EMBO Rep. e45836 (2018) doi:10.15252/embr.201845836.
9. Gao, R. et al. A highly homogeneous expansion microscopy polymer composed of tetrahedron-like monomers. bioRxiv 814111 (2019) doi:10.1101/814111.
10. Gambarotto, D. et al. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Methods 16, 71–74 (2019).
11. Pernal, S. P. et al. Differential expansion microscopy. bioRxiv 699579 (2019) doi:10.1101/699579.
12. Scheible, M. B. & Tinnefeld, P. Quantifying Expansion Microscopy with DNA Origami Expansion Nanorulers. bioRxiv (2018) doi:10.1101/265405.
13. Gao, M. et al. Expansion Stimulated Emission Depletion Microscopy (ExSTED). ACS Nano (2018) doi:10.1021/acsnano.8b00776.
14. Kim, D., Kim, T., Lee, J. & Shim, S.-H. Amplified Expansion Stimulated Emission Depletion Microscopy. ChemBioChem 20, 1260–1265 (2019).
15. Cahoon, C. K. et al. Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. 114, E6857–E6866 (2017).
16. Halpern, A. R., Alas, G. C. M., Chozinski, T. J., Paredez, A. R. & Vaughan, J. C. Hybrid Structured Illumination Expansion Microscopy Reveals Microbial Cytoskeleton Organization. ACS Nano (2017) doi:10.1021/acsnano.7b07200.
17. Wang, Y. et al. Combined expansion microscopy with structured illumination microscopy for analyzing protein complexes. Nat. Protoc. (2018) doi:10.1038/s41596-018-0023-8.
18. Shi, X. et al. Label-retention expansion microscopy. bioRxiv 687954 (2019) doi:10.1101/687954.
19. Xu, H. et al. Molecular organization of mammalian meiotic chromosome axis revealed by expansion STORM microscopy. Proc. Natl. Acad. Sci. 201902440 (2019) doi:10.1073/pnas.1902440116.
20. Zwettler, F. U. et al. Molecular resolution imaging by post-labeling expansion single molecule localization microscopy (Ex-SMLM). bioRxiv 2020.03.12.988923 (2020) doi:10.1101/2020.03.12.988923.
21. Li, R., Chen, X., Lin, Z., Wang, Y. & Sun, Y. Expansion enhanced nanoscopy. Nanoscale (2018) doi:10.1039/C8NR04267E.
22. Düring, D. N., Rocha, M. D., Dittrich, F., Gahr, M. & Hahnloser, R. H. R. Expansion Light Sheet Microscopy Resolves Subcellular Structures in Large Portions of the Songbird Brain. Front. Neuroanat. 13, (2019).
23. Gao, R. et al. Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 363, eaau8302 (2019).
24. Hörl, D. et al. BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nat. Methods (2019) doi:10.1038/s41592-019-0501-0.
25. Bürgers, J. et al. Light-sheet fluorescence expansion microscopy: fast mapping of neural circuits at super resolution. Neurophotonics 6, 015005 (2019).
26. Saka, S. K. et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol. 37, 1080–1090 (2019).
27. Sheard, T. M. D. et al. Three-Dimensional and Chemical Mapping of Intracellular Signaling Nanodomains in Health and Disease with Enhanced Expansion Microscopy. ACS Nano 13, 2143–2157 (2019).
28. Chen, F. et al. Nanoscale imaging of RNA with expansion microscopy. Nat Methods 13, 679–84 (2016).
29. Goetz, R. et al. Nanoscale imaging of bacterial infections by sphingolipid expansion microscopy. bioRxiv 2020.05.06.080663 (2020) doi:10.1101/2020.05.06.080663.
30. Karagiannis, E. D. et al. Expansion Microscopy of Lipid Membranes. bioRxiv 829903 (2019) doi:10.1101/829903.
31. Kao, P. & Nodine, M. D. Transcriptional Activation of Arabidopsis Zygotes Is Required for Initial Cell Divisions. Sci. Rep. 9, 1–11 (2019).
32. Lim, Y. et al. Mechanically resolved imaging of bacteria using expansion microscopy. PLOS Biol. 17, e3000268 (2019).
33. Yu, C.-C. (Jay) et al. Expansion microscopy of C. elegans. eLife 9, e46249 (2020).
34. Zhao, Y. et al. Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nat Biotechnol (2017) doi:10.1038/nbt.3892.